ZooGene, a DNA sequence database for calanoid copepods
and euphausiids:
an OBIS tool
for uniform standards of species identification
P.I. Ann Bucklin
Ocean Process
Analysis Laboratory
University of New
Hampshire, Durham, NH 03824
phone: (603)
862-0122 fax: (603) 862-0243
email: ann.bucklin@unh.edu
Co-P.I. Peter H.
Wiebe
Department of Biology
Woods Hole
Oceanographic Institution, Woods Hole, MA 02543
phone: (508)
268-2313 fax: (508) 457-2169 email: pwiebe@whoi.edu
Co-P.I. Bruce W.
Frost
School of
Oceanography
University of
Washington, Seattle, WA 98195
phone: (206) 543‑7186 fax: (206) 543‑6073 email: frost@ocean.washington.edu
Co-P.I. Michael J.
Fogarty
Northeast Fisheries
Science Center
National Marine
Fisheries Service, Woods Hole, MA 02543
phone: (508)
495-2386 fax: (508) 495-2258 email: fogarty@usglobec.org
NSF Grant No.
OCE-0003884
http://www.ZooGene.org
LONG-TERM GOALS
An international partnership will be established to develop
a zooplankton genomic (ZooGene) database of
DNA type sequences for calanoid copepods and euphausiids. The ZooGene database will provide molecular
systematic information that will be integrated with taxonomic and
biogeographical data [as one of the Ocean Biogeographical Information System
(OBIS) federation of databases], with molecular data [via entry-level links to
the National Center for Biotechnology Information (NCBI) molecular database
GenBank], and with environmental and oceanographic data [via the U.S. GLOBEC
Data and Information Management System].
The ZooGene database will have many potential uses, including: 1)
uniform standards of species’ identification, 2) evaluation of the taxonomic
significance of geographic variation within widespread species, 3) identification
of cryptic species, 4) accurate
estimation of species’ diversity, 5) determination of evolutionary
relationships among species, and 6) design of rapid molecularly-based species’
identification protocols.
OBJECTIVES
Objectives of the ZooGene partnership include: 1)
establishment of a nexus of taxonomic experts for calanoid copepods and
euphausiids, with focused effort on selected genera and regions; 2)
determination of mtCOI type sequences for 200 species of calanoid copepods and
86 species of euphausiids, based on verified identification of species by
taxonomic authorities; 3) design, creation, management of the ZooGene database,
a searchable web-based database integrated with the GenBank DNA sequence
database and OBIS, as a tool for uniform species identification; and 4) design
and implementation of a ZooGene curriculum module for high school students
APPROACH AND WORK PLAN
The ZooGene partnership includes four lead investigators and
thirteen expert taxonomic consultants from seven countries. Zooplankton samples are sorted from existing
archival collections, obtained in coordination with planned oceanographic
research efforts, and/or collected during National Marine Fisheries Service
field surveys. The taxonomic experts
confirm species’ identifications; DNA sequencing is done at the University of
New Hampshire and, in some cases, in other partners’ laboratories. For each species, a DNA type sequence is
determined for a portion of the mitochondrial cytochrome oxidase I (mtCOI)
gene; multiple mtCOI sequences are included as necessary to reflect
intraspecific variation. For selected
species, a portion of the slowly-evolving, nuclear small-subunit (18S) rRNA is
sequenced to examine evolutionary relationships among genera and families. The ZooGene database is designed, created,
managed, maintained, and distributed as part of the proposed work; the data
will be integrated into the Ocean Biogeographical Information System (OBIS).
WORK COMPLETED
New ZooGene collections: ZooGene partners have
collected and shipped to UNH comprehensive sets of samples for molecular
analysis using ships of opportunity.
Samples are in hand for many copepod genera, based on samples collected
and identified by Susumu Ohtsuka (Hiroshima University, Japan); David McKinnon
(Australian Institute of Marine Science); Janet Bradford-Grieve (National
Institute of Water & Atmospheric Research, New Zealand); Bruce Frost
(University of Washington); Peter Wiebe and Nancy Copley (Woods Hole
Oceanographic Institution); and Fredrica Norrbin (Tromsoe University). Euphausiid samples are in hand from Shozo
Sawamoto (Tokai University, Japan) and Jaime Färber Lorda (Centro de
Investigación Cientifica y de Educación Superior, Mexico). ZooGene samples have been collected in
association with Ecosystem Monitoring Surveys by the NOAA/NMFS Northeast
Fisheries Science Center (NEFSC), and are being used to assess spatial and
temporal variation in gene frequencies in selected taxa. New collections are now planned for the
subtropical NW Atlantic by Peter Wiebe (WHOI).
DNA type sequences:
To date, considering only the molecular analyses done in A. Bucklin’s
laboratory at UNH, the ZooGene project has determined and submitted to GenBank
150 DNA sequences for 81 species of calanoid copepods and 44 DNA sequences for
18 species of euphausiids. These
numbers are increasing rapidly with ongoing work at UNH and elswhere. MtCOI sequences have proven to be diagnostic
molecular systematic characters for both copepods and euphausiids. The sequences are also useful to reconstruct
phylogenetic relationships among congeneric species, resolve large-scale
population genetic structure and taxonomically-significant geographic
variation, and may help reveal cryptic species (Hill et al., 2001; Bucklin et
al., 2002).
RESULTS
Rapid molecular protocols for species identification:
We have designed molecular protocols based on species-specific PCR that can
discriminate cryptic, co-occurring copepod species: multiplexed SS-PCR
protocols are available for two species of Pseudocalanus (Bucklin et
al., 1999, 2001); four species of Calanus (Bucklin et al., 1999; Hill et
al., 2001; Fig. 1); and two species of Metridia (Bucklin et al, in
preparation).

Figure 1. Diagram
showing steps in the molecular identification of copepods, including A)
homogenization of the copepod in a microcentrifuge tube, B) multiplexed PCR
reaction with all species-specific reagents, and C) results of the PCR reaction
displayed on an agarose gel following electrophoresis. The species can be identified by the
migration distance of the product band in the gel. For more details, see Hill et al., 2001.
Phylogeographic analyses: Cosmopolitan species
of copepods and euphausiids have received particular attention under
ZooGene. Samples are taken across each
species’ range, in order to evaluate evidence for geographic isolation and
genetic differentiation within the species.
MtCOI has provided preliminary evidence of population genetic structure
within some geographically widespread species (Bucklin et al., 2002).
Phylogenetic reconstructions: Evolutionary relationships among congeneric copepod and
euphausiid species are typically well-resolved by mtCOI sequence
variation. MtCOI gene trees were
largely concordant with morphological phylogenetic analyses for species of both
copepods (Bucklin et al., 2002) and euphausiids (Bucklin, in preparation). Phylogenetic relationships among genera and
families of calanoid copepods and euphausiids have been examined using 18S rRNA
sequences, which provide accurate resolution (Bucklin et al., 2002, unpublished;
Grabbert et al., in preparation; Fig. 2), and are useful for comparison with
morphological analyses (Bradford-Grieve et al., unpublished).
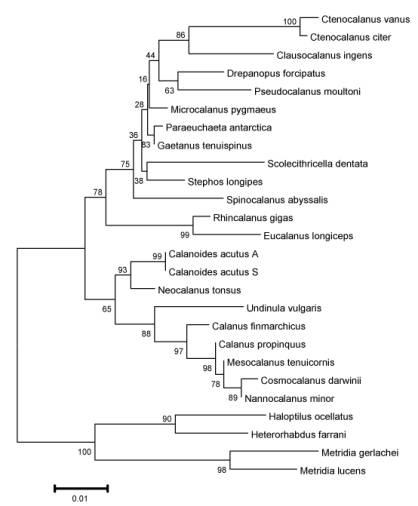
Figure 2. Gene
tree showing evolutionary relationships of selected calanoid copepod species,
based on a 660 base-pair sequence of the nuclear 18S rRNA gene. This analysis clustered together species and
genera of the same family, as expected from the alpha taxonomy of the group;
the evolutionary relationships among the families will require comparison with
morphological analyses which have not been completed yet. Data from Bucklin et al (2002) and S.
Grabbert (Univ. Oldenburg; pers. comm.).
The ZooGene database: The ZooGene database is accessible at http://www.ZooGene.org. The database includes detailed collection
information for the individual copepod or euphausiid used to determine the
mtCOI type sequence and 18S rRNA sequence.
Also included are the DNA sequences; links to the GenBank database will
be implemented shortly (via the GenBank accession number); and to environmental
data in the U.S. GLOBEC Data and Information Management System (via the
collection location). The database is
linked to and searchable from the Ocean Biogeographical Information System
(OBIS) portal/server (http://www.iobis.org/OBISPortal).
IMPACT / APPLICATIONS
We anticipate that biologists,
ecologists, and systematists will increasingly use DNA sequences as additional
characters for taxonomic identification.
The molecular information may be used to ensure uniform standards of
species identification, evaluate the taxonomic significance of geographic
variation, reveal cryptic species, accurately estimate species diversity,
determine evolutionary relationships among species, develop rapid
molecularly-based species identification protocols, and identify
morphologically-indistinguishable species at any life-stage. Despite the challenges, it is important that
ecologists and oceanographers identify species accurately. Zooplankton species are of considerable
importance in biogeochemical cycles, because of their numerical abundance,
biomass predominance, and/or critical position in coastal and ocean trophic
webs.
TRANSITIONS
Quality of Life
ZooGene will allow rapid and
accurate identification of planktonic species and estimation of marine
biodiversity, which are essential for understanding, assessing, and predicting
ecosystem health, and facilitating wise use of marine resources and sound
coastal zone planning.
Science Education and
Communication
·
We are organizing a comprehensive
systematic and molecular systematic workshop for calanoid copepods and
euphausiids to be held at the completion of ZooGene activities. The workshop will be a forum for ZooGene
partners and students, and will facilitate preparation of collaborative manuscripts
and renewal proposals.
·
At the University of
Washington, a graduate student (Mikelle Rasmussen) is nearing completion of the
Masters’ degree: Genetic variation
among subspecies of the planktonic copepod Calanus pacificus: A ZooGene
Application. University of Washington, Seattle.
·
An international student
exchange was conducted between UNH and the Alfred Wegener Institute in
Germany. Sabine Grabbert, a PhD student
of Dr. Sigrid Schiel, spent 6 months in A. Bucklin’s laboratory at UNH, completing
genetic analysis of copepods that will be included in her doctoral dissertation
for the University of Oldenburg (Germany).
·
A ZooGene molecular kit and
curriculum module were designed and implemented for use in New Hampshire high
schools. An informative brochure for
teachers was produced, and a one-day work-shop for 5 teachers was held in
August, 2000. A NH Sea Grant Marine
Docent is taking the lead in this effort, which is now being tested in local
high schools.
·
A project web site, http://www.ZooGene.org, has
been established. The web site provides
general information on the partnership, as well as technical information to
guide colleagues in collecting and preserving zooplankton for molecular
analysis.
REFERENCES
Bucklin, A., R.S. Hill, M. Guarnieri, A.M. Bentley, and S.
Kaartvedt (1999) Taxonomic and
systematic assessment of planktonic copepods using mitochondrial COI sequence
variation and competitive, species-specific PCR. Hydrobiol. 401:239-254.
Bucklin, A., M. Guarnieri, D.J. McGillicuddy, and R.S. Hill
(2001) Spring evolution of Pseudocalanus
spp. abundance on Georges Bank based on molecular discrimination of P.
moultoni and P. newmani.
Deep-Sea Res. 48: 589-609.
PUBLICATIONS
Bucklin, A. (2000)
Methods for Population Genetic Analysis of Zooplankton. Chapter 11 in: The Zooplankton
Methodology Manual, International Council for the Exploration of the Sea.
Academic press, London. Pp. 533-570.
Hill, R.S., L.D. Allen, and A. Bucklin (2001) Multiplexed species-specific PCR protocol to
discriminate four N. Atlantic Calanus species, with a mtCOI gene tree
for ten Calanus species. Marine
Biology 139: 279-287.
Bucklin, A., P.H. Wiebe, S. B. Smolenack, N.J. Copley, and
M.E. Clarke (2002) Integrated
biochemical, molecular genetic, and bioacoustical analysis of mesoscale
variability of the euphausiid Nematoscelis difficilis in the California
Current. Deep-Sea Res. 49: 437-462.
Bucklin, A., B.W.
Frost, J. Bradford-Grieve, L.D. Allen
and N.J. Copley (2003) Molecular systematic assessment of thirty-four calanoid
copepod species of the Calanidae and Clausocalanidae using DNA sequences of
mtCOI and nuclear 18S rRNA. Mar. Biol.
142: 333-343.
Caudill, C.C. and A. Bucklin. Population genetic diversity
of Acartia tonsa: evidence for cryptic speciation. Hydrobiol. (In revision)

